
检验医学 ›› 2023, Vol. 38 ›› Issue (9): 818-824.DOI: 10.3969/j.issn.1673-8640.2023.09.003
收稿日期:2022-05-24
修回日期:2022-12-04
出版日期:2023-09-30
发布日期:2023-11-29
通讯作者:
赵晓君,E-mail:zhaoxiaojun@sccl.org.cn。
作者简介:孙传玉,男,1984年生,博士,副主任医师,主要从事前列腺癌相关生物标记物筛选研究。
SUN Chuanyu1, ZHAO Xiaojun2( ), GE Shengyang1, ZHANG Yang3
), GE Shengyang1, ZHANG Yang3
Received:2022-05-24
Revised:2022-12-04
Online:2023-09-30
Published:2023-11-29
摘要:
目的 对相关研究发现的46个前列腺癌差异表达蛋白的上游转录调控机制进行分析和探讨。方法 采用GeneGo MetaCore软件和IPA软件对46个差异蛋白进行生物信息学分析,明确这些差异蛋白的转录因子调控网络。采用GEPIA2数据库分析鉴定出的转录因子的表达量及其对前列腺癌患者生存情况的影响。结果 共鉴定出20个转录调控网络。按差异表达蛋白的富集度进行排序,筛选出居前6位的转录调控网络,分别由特化蛋白1(SP1)、p53、阴阳1(YY1)、雄激素受体(AR)、c-Myc和Slug 6个转录因子调控。Kaplan-Meier生存曲线分析结果显示,高表达p53的前列腺癌患者无病生存期和总生存期均长于低表达p53的患者(P值分别为0.047、0.019),低表达AR的前列腺癌患者无病生存期长于高表达AR的患者(P=0.023),其余4个转录因子对前列腺癌患者预后无影响(P>0.05)。IPA软件分析结果显示,SP1、p53、YY1、AR和c-Myc由表没食子儿茶素没食子酸酯(EGCG)、蛋白激酶B(AKT)1、成纤维细胞生长因子2(FGF2)、β-雌二醇调控。结论 通过分析46个前列腺癌差异表达蛋白的转录调控网络,发现6个与前列腺癌相关的转录因子和4个上游调节分子,可为前列腺癌早期筛查和预后评估提供新的靶标。
中图分类号:
孙传玉, 赵晓君, 葛圣阳, 张扬. 前列腺癌进展相关转录因子研究[J]. 检验医学, 2023, 38(9): 818-824.
SUN Chuanyu, ZHAO Xiaojun, GE Shengyang, ZHANG Yang. Transcription factors in prostate cancer progression[J]. Laboratory Medicine, 2023, 38(9): 818-824.
| 蛋白编号 | 蛋白名称 | 肽段(95%) | 116∶114① |
|---|---|---|---|
| P62241 | 40S核糖体蛋白S8 | 2 | 1.53 |
| P31943 | 异质核糖核蛋白H | 3 | 2.17 |
| P11021 | GRP78② | 27 | 2.21 |
| P09669 | 细胞色素C氧化酶6抗体 | 2 | 2.38 |
| P10606 | 细胞色素c氧化酶亚单位5B,线粒体 | 7 | 2.70 |
| P06576 | ATP合酶β亚单位,线粒体 | 13 | 2.83 |
| P38646 | 应激70蛋白,线粒体 | 11 | 2.91 |
| P21810 | 二聚糖 | 19 | 2.97 |
| P49411 | 延伸因子Tu,线粒体 | 6 | 3.02 |
| P22314 | 泛素样修饰活化酶1 | 5 | 3.34 |
| P13667 | 蛋白二硫化物异构酶A4 | 5 | 3.77 |
| Q5SSJ5 | 异染色质蛋白1-结合蛋白3 | 5 | 4.06 |
| P55327 | 肿瘤蛋白D52 | 3 | 4.25 |
| Q99623 | 抗增殖蛋白2 | 3 | 4.57 |
| P07237 | 蛋白质二硫化物异构酶 | 17 | 4.70 |
| Q9NZN4 | EH结构域蛋白2 | 2 | 5.55 |
| P10809 | HSP60③,线粒体 | 25 | 5.97 |
| P06748 | 核磷胺 | 10 | 7.94 |
| Q15063 | 成骨细胞特异性因子2 | 13 | 9.12 |
| Q00796 | 山梨醇脱氢酶 | 9 | 9.82 |
表1 表达上调的前列腺癌差异表达蛋白[10]
| 蛋白编号 | 蛋白名称 | 肽段(95%) | 116∶114① |
|---|---|---|---|
| P62241 | 40S核糖体蛋白S8 | 2 | 1.53 |
| P31943 | 异质核糖核蛋白H | 3 | 2.17 |
| P11021 | GRP78② | 27 | 2.21 |
| P09669 | 细胞色素C氧化酶6抗体 | 2 | 2.38 |
| P10606 | 细胞色素c氧化酶亚单位5B,线粒体 | 7 | 2.70 |
| P06576 | ATP合酶β亚单位,线粒体 | 13 | 2.83 |
| P38646 | 应激70蛋白,线粒体 | 11 | 2.91 |
| P21810 | 二聚糖 | 19 | 2.97 |
| P49411 | 延伸因子Tu,线粒体 | 6 | 3.02 |
| P22314 | 泛素样修饰活化酶1 | 5 | 3.34 |
| P13667 | 蛋白二硫化物异构酶A4 | 5 | 3.77 |
| Q5SSJ5 | 异染色质蛋白1-结合蛋白3 | 5 | 4.06 |
| P55327 | 肿瘤蛋白D52 | 3 | 4.25 |
| Q99623 | 抗增殖蛋白2 | 3 | 4.57 |
| P07237 | 蛋白质二硫化物异构酶 | 17 | 4.70 |
| Q9NZN4 | EH结构域蛋白2 | 2 | 5.55 |
| P10809 | HSP60③,线粒体 | 25 | 5.97 |
| P06748 | 核磷胺 | 10 | 7.94 |
| Q15063 | 成骨细胞特异性因子2 | 13 | 9.12 |
| Q00796 | 山梨醇脱氢酶 | 9 | 9.82 |
| 蛋白编号 | 蛋白名称 | 肽段(95%) | 116∶114 |
|---|---|---|---|
| P17661 | 结蛋白 | 62 | 0.03 |
| O95810 | 血清剥夺反应蛋白3 | 3 | 0.13 |
| Q9BX66 | SORBS1 | 18 | 0.18 |
| P08670 | 波形蛋白 | 65 | 0.19 |
| P51911 | 钙调蛋白-1 | 43 | 0.20 |
| P04792 | β-1热休克蛋白 | 15 | 0.22 |
| P63267 | γ-肠平滑肌肌动蛋白 | 191 | 0.24 |
| Q969G5 | 蛋白激酶C δ结合蛋白 | 6 | 0.24 |
| P12109 | 胶原蛋白α-1(Ⅵ)链 | 19 | 0.25 |
| P11047 | γ-1层黏连蛋白 | 9 | 0.28 |
| O94875 | SORBS2 | 2 | 0.32 |
| P24821 | 肌腱蛋白 | 7 | 0.35 |
| Q15942 | Zyxin | 4 | 0.35 |
| Q9Y490 | Talin-1 | 21 | 0.36 |
| P02452 | α-1(Ⅰ)链胶原蛋白 | 92 | 0.38 |
| P18206 | 黏着斑蛋白 | 15 | 0.39 |
| P20774 | Mimecan | 6 | 0.40 |
| Q93052 | 脂肪瘤相关抗体 | 12 | 0.41 |
| Q09666 | 神经细胞分化相关蛋白 | 62 | 0.42 |
| P07585 | 核心蛋白聚糖 | 12 | 0.43 |
| P98160 | 基底膜特异性硫酸乙酰肝素蛋白聚糖核心蛋白 | 17 | 0.44 |
| P30086 | 磷脂酰乙醇胺结合蛋白1 | 7 | 0.47 |
| P06396 | 凝溶胶蛋白 | 20 | 0.49 |
| P12111 | 胶原蛋白α-3(Ⅵ)链 | 63 | 0.50 |
| P01834 | Ig κ链C区 | 4 | 0.54 |
| P10909 | 簇集蛋白 | 3 | 0.54 |
表2 表达下调的前列腺癌差异表达蛋白[10]
| 蛋白编号 | 蛋白名称 | 肽段(95%) | 116∶114 |
|---|---|---|---|
| P17661 | 结蛋白 | 62 | 0.03 |
| O95810 | 血清剥夺反应蛋白3 | 3 | 0.13 |
| Q9BX66 | SORBS1 | 18 | 0.18 |
| P08670 | 波形蛋白 | 65 | 0.19 |
| P51911 | 钙调蛋白-1 | 43 | 0.20 |
| P04792 | β-1热休克蛋白 | 15 | 0.22 |
| P63267 | γ-肠平滑肌肌动蛋白 | 191 | 0.24 |
| Q969G5 | 蛋白激酶C δ结合蛋白 | 6 | 0.24 |
| P12109 | 胶原蛋白α-1(Ⅵ)链 | 19 | 0.25 |
| P11047 | γ-1层黏连蛋白 | 9 | 0.28 |
| O94875 | SORBS2 | 2 | 0.32 |
| P24821 | 肌腱蛋白 | 7 | 0.35 |
| Q15942 | Zyxin | 4 | 0.35 |
| Q9Y490 | Talin-1 | 21 | 0.36 |
| P02452 | α-1(Ⅰ)链胶原蛋白 | 92 | 0.38 |
| P18206 | 黏着斑蛋白 | 15 | 0.39 |
| P20774 | Mimecan | 6 | 0.40 |
| Q93052 | 脂肪瘤相关抗体 | 12 | 0.41 |
| Q09666 | 神经细胞分化相关蛋白 | 62 | 0.42 |
| P07585 | 核心蛋白聚糖 | 12 | 0.43 |
| P98160 | 基底膜特异性硫酸乙酰肝素蛋白聚糖核心蛋白 | 17 | 0.44 |
| P30086 | 磷脂酰乙醇胺结合蛋白1 | 7 | 0.47 |
| P06396 | 凝溶胶蛋白 | 20 | 0.49 |
| P12111 | 胶原蛋白α-3(Ⅵ)链 | 63 | 0.50 |
| P01834 | Ig κ链C区 | 4 | 0.54 |
| P10909 | 簇集蛋白 | 3 | 0.54 |
| 序号 | 转录因子 | GO富集生物过程 | 总节点数/个 | 根节点数/个 | P值 |
|---|---|---|---|---|---|
| 1 | SP1 | 细胞组分转移、细胞黏附调控、生物特性调控 | 19 | 18 | 9.23×10-57 |
| 2 | p53 | 细胞凋亡的负调控 | 11 | 10 | 4.70×10-31 |
| 3 | YY1 | 细胞、器官发育、生物过程的负调控,细胞黏附的负调控 | 10 | 9 | 6.55×10-28 |
| 4 | AR | 细胞凋亡负调控,抑制细胞增殖 | 8 | 7 | 1.14×10-21 |
| 5 | c-Myc | 细胞凋亡负调控,Caspase②活性调节 | 8 | 7 | 1.14×10-21 |
| 6 | Slug | 生物过程的负调节,参与氧化应激反应 | 48 | 28 | 1.12×10-78 |
表3 SP1、p53、YY1、AR、c-Myc和Slug的GO①富集分析
| 序号 | 转录因子 | GO富集生物过程 | 总节点数/个 | 根节点数/个 | P值 |
|---|---|---|---|---|---|
| 1 | SP1 | 细胞组分转移、细胞黏附调控、生物特性调控 | 19 | 18 | 9.23×10-57 |
| 2 | p53 | 细胞凋亡的负调控 | 11 | 10 | 4.70×10-31 |
| 3 | YY1 | 细胞、器官发育、生物过程的负调控,细胞黏附的负调控 | 10 | 9 | 6.55×10-28 |
| 4 | AR | 细胞凋亡负调控,抑制细胞增殖 | 8 | 7 | 1.14×10-21 |
| 5 | c-Myc | 细胞凋亡负调控,Caspase②活性调节 | 8 | 7 | 1.14×10-21 |
| 6 | Slug | 生物过程的负调节,参与氧化应激反应 | 48 | 28 | 1.12×10-78 |
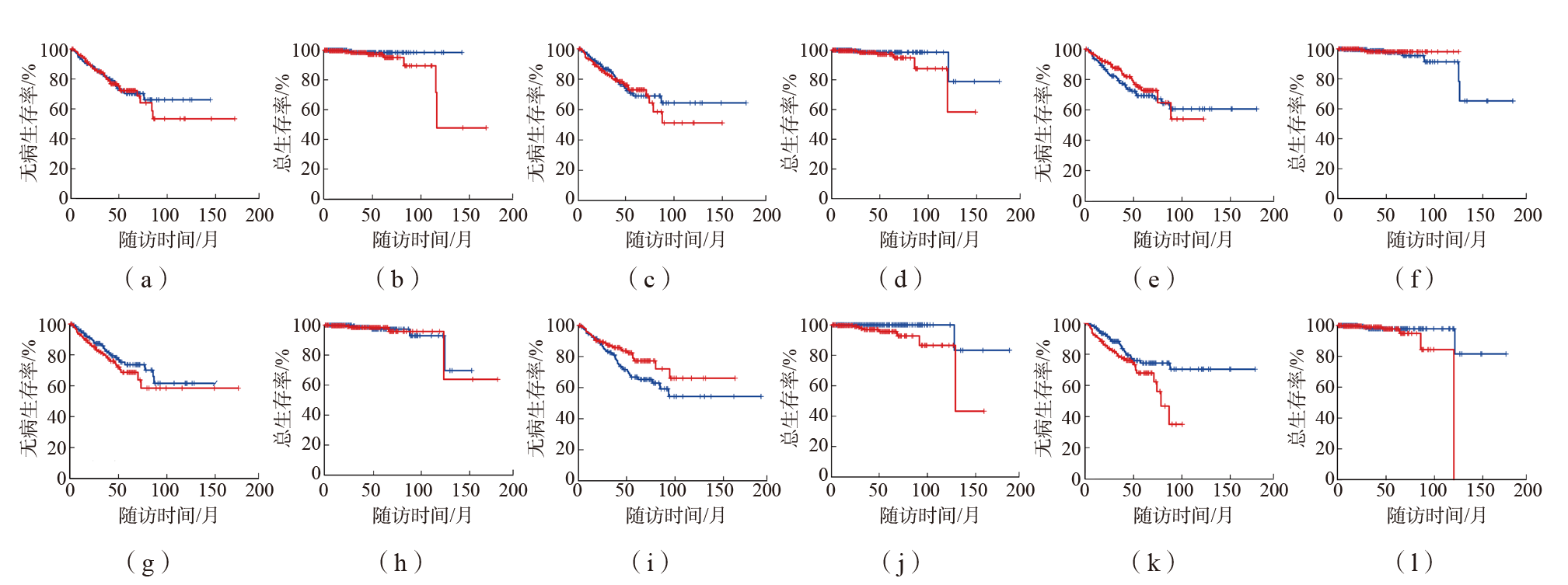
图2 c-Myc、YY1、Slug、SP1、p53、AR对前列腺癌患者无病生存期和总生存期的影响 (a)c-Myc对前列腺癌患者无病生存期的影响; 低表达c-Myc, 高表达c-Myc;(b)c-Myc对前列腺癌患者总生存期的影响; 低表达c-Myc, 高表达c-Myc;(c)YY1对前列腺癌患者无病生存期的影响; 低表达YY1, 高表达YY1;(d)YY1对前列腺癌患者总生存期的影响; 低表达YY1, 高表达YY1;(e)Slug对前列腺癌患者无病生存期的影响; 低表达Slug, 高表达Slug;(f)Slug对前列腺癌患者总生存期的影响; 低表达Slug, 高表达Slug;(g)SP1对前列腺癌患者无病生存期的影响; 低表达SP1, 高表达SP1;(h)SP1对前列腺癌患者总生存期的影响; 低表达SP1, 高表达SP1;(i)p53对前列腺癌患者无病生存期的影响; 低表达p53, 高表达p53;(j)p53对前列腺癌患者总生存期的影响; 低表达p53, 高表达p53;(k)AR对前列腺癌患者无病生存期的影响; 低表达AR, 高表达AR;(l)AR对前列腺癌患者总生存期的影响; 低表达AR, 高表达AR。
| [1] |
SIEGEL R L, MILLER K D, FUCHS H E, et al. Cancer statistics,2022[J]. CA Cancer J Clin, 2022, 72(1):7-33.
DOI URL |
| [2] |
GE S, MI Y, ZHAO X, et al. Characterization and validation of long noncoding RNAs as new candidates in prostate cancer[J]. Cancer Cell Int, 2020, 20(1):531.
DOI PMID |
| [3] |
SUNG H, FERLAY J, SIEGEL R L, et al. Global cancer statistics 2020:globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3):209-249.
DOI URL |
| [4] |
CHEN Q, FU L, HU J, et al. Silencing of PSMC2 inhibits development and metastasis of prostate cancer through regulating proliferation,apoptosis and migration[J]. Cancer Cell Int, 2021, 21(1):235.
DOI |
| [5] |
AFFRONTI H C, ROWSAM A M, PELLERITE A J, et al. Pharmacological polyamine catabolism upregulation with methionine salvage pathway inhibition as an effective prostate cancer therapy[J]. Nat Commun, 2020, 11(1):52.
DOI PMID |
| [6] |
LONG P, ZHANG L, HUANG B, et al. Integrating genome sequence and structural data for statistical learning to predict transcription factor binding sites[J]. Nucleic Acids Res, 2020, 48(22):12604-12617.
DOI PMID |
| [7] |
YANG M, HUANG W, SUN Y, et al. Prognosis and modulation mechanisms of COMMD6 in human tumours based on expression profiling and comprehensive bioinformatics analysis[J]. Br J Cancer, 2019, 121(8):699-709.
DOI |
| [8] |
GONDANE A, GIRMAY S, HELEVÄ A, et al. O-GlcNAc transferase couples MRE11 to transcriptionally active chromatin to suppress DNA damage[J]. J Biomed Sci, 2022, 29(1):13.
DOI PMID |
| [9] |
LOU S, LI T, KONG X, et al. TopicNet:a framework for measuring transcriptional regulatory network change[J]. Bioinformatics, 2020, 36(Suppl 1):i474-i481.
DOI URL |
| [10] |
SUN C Y, MI Y Y, GE S Y, et al. Tumor- and osteoblast-derived periostin in prostate cancer bone metastases[J]. Front Oncol, 2022, 11:795712.
DOI URL |
| [11] |
SUN C, SONG C, MA Z, et al. Periostin identified as a potential biomarker of prostate cancer by iTRAQ-proteomics analysis of prostate biopsy[J]. Proteome Sci, 2011, 9:22.
DOI PMID |
| [12] |
SUN Y, XU K, HE M, et al. Overexpression of glypican 5(GPC5) inhibits prostate cancer cell proliferation and invasion via suppressing Sp1-mediated EMT and activation of Wnt/β-catenin signaling[J]. Oncol Res, 2018, 26(4):565-572.
DOI URL |
| [13] | KANG D, ZUO W, WU Q, et al. Inhibition of specificity protein 1 is involved in phloretin-induced suppression of prostate cancer[J]. Biomed Res Int, 2020, 2020:1358674. |
| [14] |
HAN D S, LEE E O. Sp1 plays a key role in vasculogenic mimicry of human prostate cancer cells[J]. Int J Mol Sci, 2022, 23(3):1321.
DOI URL |
| [15] |
TAKAYAMA K I, SUZUKI T, FUJIMURA T, et al. Association of USP 10 with G3BP2 inhibits p53 signaling and contributes to poor outcome in prostate cancer[J]. Mol Cancer Res, 2018, 16(5):846-856.
DOI URL |
| [16] |
ASHIKARI D, TAKAYAMA K, TANAKA T, et al. Androgen induces G3BP2 and SUMO-mediated p53 nuclear export in prostate cancer[J]. Oncogene, 2017, 36(45):6272-6281.
DOI PMID |
| [17] |
PASCAL L E, WANG Y, ZHONG M, et al. EAF2 and p53 co-regulate STAT3 activation in prostate cancer[J]. Neoplasia, 2018, 20(4):351-363.
DOI PMID |
| [18] |
SARVAGALLA S, KOLAPALLI S P, VALLABHAPURAPU S. The two sides of YY1 in cancer:a friend and a foe[J]. Front Oncol, 2019, 9:1230.
DOI URL |
| [19] |
XU C, TSAI Y H, GALBO P M, et al. Cistrome analysis of YY1 uncovers a regulatory axis of YY1:BRD2/4-PFKP during tumorigenesis of advanced prostate cancer[J]. Nucleic Acids Res, 2021, 49(9):4971-4988.
DOI PMID |
| [20] |
CAMACHO-MOCTEZUMA B, QUEVEDO-CASTILLO M, MELENDEZ-ZAJGLA J, et al. YY1 negatively regulates the XAF1 gene expression in prostate cancer[J]. Biochem Biophys Res Commun, 2019, 508(3):973-979.
DOI URL |
| [21] |
LU C, BROWN L C, ANTONARAKIS E S, et al. Androgen receptor variant-driven prostate cancer Ⅱ:advances in laboratory investigations[J]. Prostate Cancer Prostatic Dis, 2020, 23(3):381-397.
DOI |
| [22] |
FRAME F M, MAITLAND N J. Epigenetic control of gene expression in the normal and malignant human prostate:a rapid response which promotes therapeutic resistance[J]. Int J Mol Sci, 2019, 20(10):2437.
DOI URL |
| [23] |
NANDA J S, AWADALLAH W N, KOHRT S E, et al. Increased nuclear factor I/B expression in prostate cancer correlates with AR expression[J]. Prostate, 2020, 80(13):1058-1070.
DOI PMID |
| [24] |
SHORNING B Y, DASS M S, SMALLEY M J, et al. The PI3K-AKT-mTOR pathway and prostate cancer:at the crossroads of AR,MAPK,and WNT signaling[J]. Int J Mol Sci, 2020, 21(12):4507.
DOI URL |
| [25] |
LABBÉ D P, ZADRA G, YANG M, et al. High-fat diet fuels prostate cancer progression by rewiring the metabolome and amplifying the MYC program[J]. Nat Commun, 2019, 10(1):4358.
DOI PMID |
| [26] |
LABBÉ D P, BROWN M. Transcriptional regulation in prostate cancer[J]. Cold Spring Harb Perspect Med, 2018, 8(11):a030437.
DOI URL |
| [27] |
MICKOVA A, KHARAISHVILI G, KURFURSTOVA D, et al. Skp2 and Slug are coexpressed in aggressive prostate cancer and inhibited by neddylation blockade[J]. Int J Mol Sci, 2021, 22(6):2844.
DOI URL |
| [28] |
IWASAKI K, NINOMIYA R, SHIN T, et al. Chronic hypoxia-induced slug promotes invasive behavior of prostate cancer cells by activating expression of ephrin-B1[J]. Cancer Sci, 2018, 109(10):3159-3170.
DOI URL |
| [29] |
MARCHETTI C, GAVAZZO P, BURLANDO B. Epigallocatechin-3-gallate mobilizes intracellular Ca2+ in prostate cancer cells through combined Ca2+ entry and Ca2+-induced Ca2+ release[J]. Life Sci, 2020, 258:118232.
DOI URL |
| [30] |
KHODAYARI MOEZ E, PYNE S, DINU I. Association between bivariate expression of key oncogenes and metabolic phenotypes of patients with prostate cancer[J]. Comput Biol Med, 2018, 103:55-63.
DOI PMID |
| [31] |
VASILEIOU C, BEFANI C, DIMAS K, et al. FGF-2 and HGF reverse abiraterone's effect οn intracellular levels of DHT in androgen-dependent and androgen independent prostate cancer cell lines[J]. J BUON, 2020, 25(2):1141-1147.
PMID |
| [32] |
DI ZAZZO E, GALASSO G, GIOVANNELLI P, et al. Estrogens and their receptors in prostate cancer:therapeutic implications[J]. Front Oncol, 2018, 8:2.
DOI URL |
| [1] | 嵇冉, 李娜, 张玉, 刘一鸣. 颅脑外伤患儿血清FoxO3a和Cav-1水平变化及其临床意义[J]. 检验医学, 2024, 39(8): 728-732. |
| [2] | 方丹东, 程岗, 黄伟, 刘晓楠, 毛尖, 侯保森, 刘十岷. 脑胶质瘤组织lncRNA SOX21-AS1、miR-875-5p表达及其与患者预后的关系[J]. 检验医学, 2024, 39(3): 209-214. |
| [3] | 诸宏伟, 张雪灵, 王美娣, 郑迎娟. LZTR1基因Arg284His变异致Noonan综合征10型病例报道和遗传学分析[J]. 检验医学, 2024, 39(2): 120-125. |
| [4] | 邹琛, 徐润灏, 丁毅, 张洁, 翁文浩, 王震华, 曹芸. 基于ProteomeXchange数据库建立结直肠癌筛查模型[J]. 检验医学, 2024, 39(12): 1181-1189. |
| [5] | 张勤, 姚瀚鑫, 王伯玉, 巨昕薇, 续薇. t-PSA及其衍生指标PHI和PI-RADS评分对前列腺癌诊断效能的评价[J]. 检验医学, 2024, 39(11): 1084-1090. |
| [6] | 彭伟, 李运改, 许静, 刘华, 杨翠霞, 沈云岳. 血清炎症因子联合PSA、f-PSA可辅助诊断前列腺癌[J]. 检验医学, 2023, 38(9): 849-854. |
| [7] | 吴炯, 胡嘉华, 施美芳, 刘涛, 戴洁, 卢忻怡, 邹政. 前列腺癌生物标志物研究进展[J]. 检验医学, 2023, 38(2): 190-195. |
| [8] | 白伟, 王斌, 郭甲瑞, 朱光谱, 朱丽艳. 骨肉瘤组织PBX1表达及其对患者预后的影响[J]. 检验医学, 2023, 38(10): 926-929. |
| [9] | 叶敬文, 沈云岳, 刘鷖雯, 何怡青, 杜艳, 张国良, 高锋, 杨翠霞. MAPK/ERK信号通路在逆转乳腺癌内分泌治疗耐药中的作用[J]. 检验医学, 2022, 37(4): 342-348. |
| [10] | 王琳琳, 许丽丽, 范君, 钱余. 氧化应激标志物与前列腺增生和前列腺癌的相关性[J]. 检验医学, 2022, 37(12): 1135-1140. |
| [11] | 权衡, 朱婧, 廖焕金, 伍均. p2PSA、p2PSA%、PHI在前列腺癌辅助诊断中的价值[J]. 检验医学, 2021, 36(7): 705-709. |
| [12] | 黄晓峰, 范雪明, 姚天悦, 袁文华, 赵智赟, 宋云霄. 外周血红细胞平均分布宽度与前列腺癌的相关性研究[J]. 检验医学, 2021, 36(6): 590-595. |
| [13] | 郦俊生, 宋云霄, 程捷. 治疗前PLR、NLR和LMR在预测前列腺癌预后中的价值[J]. 检验医学, 2021, 36(6): 631-636. |
| [14] | 俞莹, 李建杰, 李汉华, 黄娟, 翁文浩, 陈学飞. miR-4429通过MTDH抑制前列腺癌细胞增殖、迁移与侵袭[J]. 检验医学, 2021, 36(12): 1267-1273. |
| [15] | 李婵婵, 王茹, 罗文英. 铜绿假单胞菌转录因子PsrA全局调控研究进展[J]. 检验医学, 2020, 35(8): 832-836. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||